Abstract
Background and aims. Data collection in clinical trials is becoming complex, with huge amount of clinical and biological variables that need to be recorded, verified and analyzed to confirm obvious and identify unexpected associations. Outside the medical field, data warehouses (DWs) are widely employed to achieve these objectives. To verify whether DWs might be useful for data quality and association analysis, a team of clinicians, biologists, statisticians and biomedical engineers developed DELPHI ("Data ELaboration to Predict Hypothetical assocIations"). The tool has been suited to a large clinical trial from the Fondazione Italiana Linfomi (FIL), including all the clinical, biological and mutational features collected both in the trial and in the ancillary studies. Herein we present the first results, related to baseline variables: 1) creation of a broad DW (including 350 variables for 300 patients, thus > 105data); 2) data harmonization and quality control; 3) discovery of unexpected associations and (in a later phase) outcome correlations; 4) results visualization.
Methods. Data were retrieved from electronic case reporting forms (eCRF) of the phase III, multicenter "FIL-MCL0208" trial (NCT02354313) for younger, untreated mantle cell lymphoma patients [Cortelazzo, EHA 2015], now at the second interim analysis. DW was created with Mysql®, a relational database management system, already used in many apps. DW filling, data cleaning and statistical analysis were performed via Matlab®. For this preliminary analysis 62 baseline features from 300 subjects were organized in groups of demographics, clinical, laboratory, pathological, minimal residual disease, gene expression profiling (GEP) and mutational variables. A first data quality control was performed. Secondly, a team of 3 lymphoma experts claimed the expected associations among about 1860 couples of features conveniently categorized. The original series was randomly divided in two subsets (200 vs 100 subjects): the first used for discovery and the second as a validation set. For each subset, couples were screened by Chi-square and Fisher's test as appropriate (p<0.05), then Cramer's V coefficients were calculated to assess the statistical strength of association. Couples from discovery set confirmed by validation-set were selected. Hence, selected couples were overlapped to the previously declared "clinical expectations" to extrapolate the unexpected ones, suggested by the analysis. Data visualization was performed via Circos [Krzywinski, Gen Res 2009].
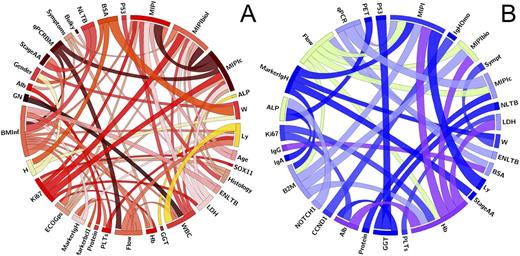
Results. Quality Control: DELPHI allowed cross comparison analysis and detected many incongruities in eCRF, prompting request for queries to clinical centers, which allowed data corrections - e.g. data quality increase has been recorded for classical laboratory (+7%), biological (+40%) and clinical data (up to 99.6% of correctness). Testing associations: DELPHI identified 231/1860 (12%) associations in the discovery set, of whom 64% (149/231) were confirmed in the validation set. Among these mismatches, main contribution is imputed to associations between clinical and laboratory data with baseline tumor burden by quantitative PCR, qPCR, (n=21) and mutational data (n=11). The clinical team classified as "expected" 242/1860 variables matches (13%), 54% of whom were confirmed by DELPHI (Figure 1A). The thickest ribbons have V>0.5: among these, bone marrow tumor invasion (BMinf) by immunohistochemistry with nodal (NLTB) and extra-nodal (ENLTB) tumor burden at CT scan; Lymphocytes with BMinf by Flow cytometry (Flow). TP53 mutations and blastoid histology with a V=0.39. Finally, discovery of novel associations is shown in Figure 1B. 54 of 1860 (3%) matches were identified by DELPHI as statistically significant unexpected associations: the thickest ribbons have V>0.35. Among these, MIPI, albumin and LDH with Hemoglobin; BMinf by qPCR with MIPIc. Moreover, TP53 mutations and GammaGT (V=0.34), as well as NOTCH1 mutations with ALkaline-Phosphatase (V=0.35) and MIPIb with Beta2 Microglobulin, B2M (V=0.33).
Discussion. DELPHI DW is a powerful tool that identifies novel putative associations between clinical and biological features. In order to consider confounding interactions and multiple comparison issues, every association needs further validation on independent data series. The association of baseline data with post-treatment outcomes is ongoing.
Galimberti: Novartis: Speakers Bureau; Incyte: Speakers Bureau; Bristol-Myers Squibb: Speakers Bureau; Pfizer: Speakers Bureau. Gaidano: AbbVie: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Gilead: Consultancy, Honoraria; Roche: Consultancy, Honoraria. Boccadoro: AbbVie: Honoraria; Amgen: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Novartis: Honoraria, Research Funding; Bristol-Myers Squibb: Honoraria, Research Funding; Sanofi: Honoraria, Research Funding; Celgene: Honoraria, Research Funding; Mundipharma: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal